Enabling Detailed, Biophysics-Based Skeletal Muscle Models on HPC Systems
 (opens in new tab)
(opens in new tab)
Venue. Frontiers in Physiology (2018)
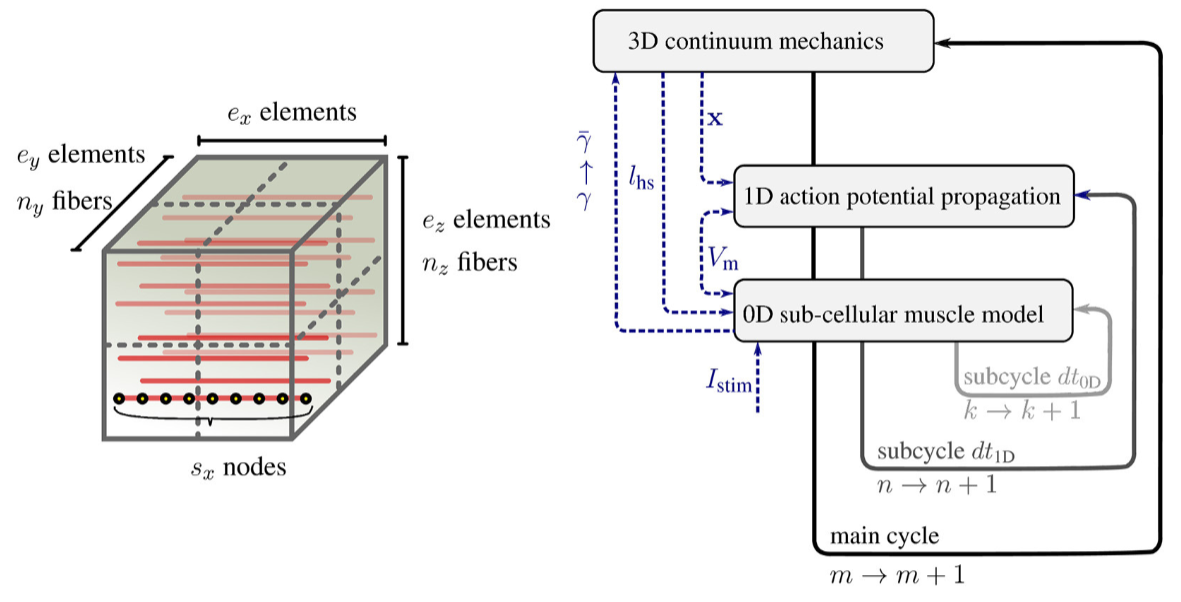
Abstract. Realistic simulations of detailed, biophysics-based, multi-scale models often require very high resolution and, thus, large-scale compute facilities. Existing simulation environments, especially for biomedical applications, are typically designed to allow for high flexibility and generality in model development. Flexibility and model development, however, are often a limiting factor for large-scale simulations. Therefore, new models are typically tested and run on small-scale compute facilities. By using a detailed biophysics-based, chemo-electromechanical skeletal muscle model and the international open-source software library OpenCMISS as an example, we present an approach to upgrade an existing muscle simulation framework from a moderately parallel version toward a massively parallel one that scales both in terms of problem size and in terms of the number of parallel processes. For this purpose, we investigate different modeling, algorithmic and implementational aspects. We present improvements addressing both numerical and parallel scalability. In addition, our approach includes a novel visualization environment which is based on the MegaMol framework and is capable of handling large amounts of simulated data. We present the results of a number of scaling studies at the Tier-1 supercomputer HazelHen at the High Performance Computing Center Stuttgart (HLRS). We improve the overall runtime by a factor of up to 2.6 and achieve good scalability on up to 768 cores.
Link to this page:

